Quickstart#
import pytometry as pm
import readfcs
import anndata
Read fcs file example from the readfcs package.
path_data = readfcs.datasets.example()
adata = pm.io.read_fcs(path_data)
assert isinstance(adata, anndata._core.anndata.AnnData)
pm.pp.split_signal(adata, var_key="channel")
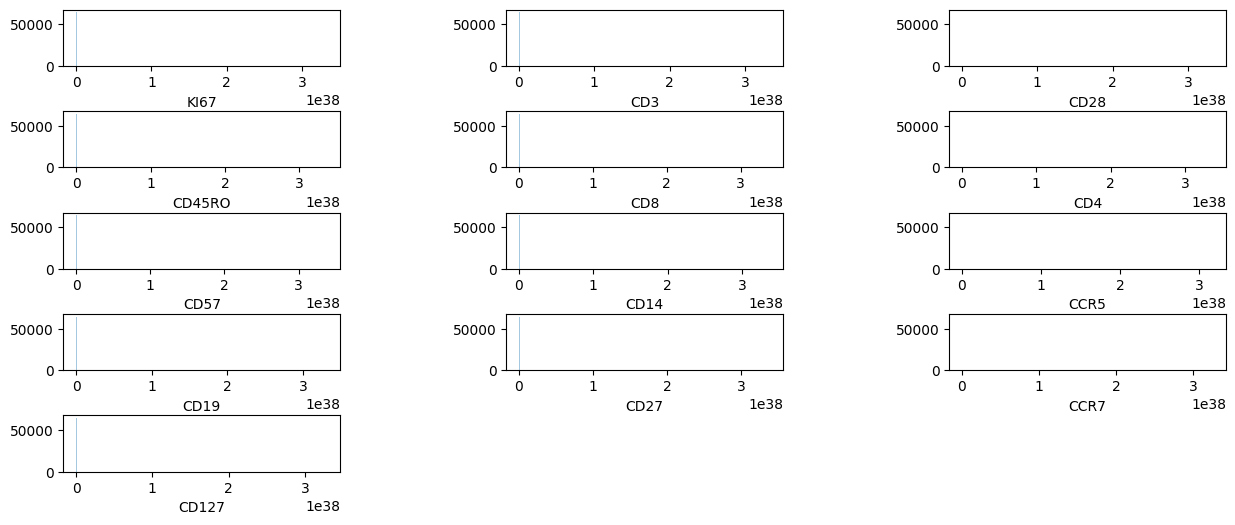
pm.pl.plotdata(adata)
/home/runner/work/pytometry/pytometry/.nox/build-3-9/lib/python3.9/site-packages/pytometry/plotting/_histogram.py:83: UserWarning:
`distplot` is a deprecated function and will be removed in seaborn v0.14.0.
Please adapt your code to use either `displot` (a figure-level function with
similar flexibility) or `histplot` (an axes-level function for histograms).
For a guide to updating your code to use the new functions, please see
https://gist.github.com/mwaskom/de44147ed2974457ad6372750bbe5751
sns.distplot(
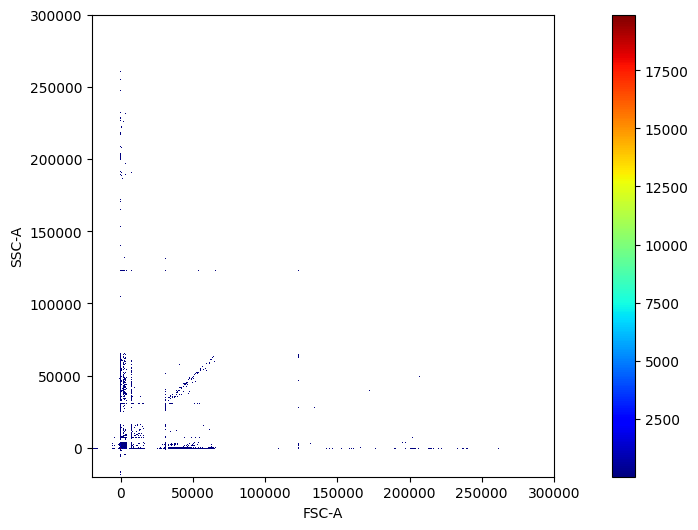
pm.pl.scatter_density(adata)
pm.pp.compensate(adata)
5499 NaN values found after compensation. Please adjust compensation matrix.
/home/runner/work/pytometry/pytometry/.nox/build-3-9/lib/python3.9/site-packages/pytometry/preprocessing/_process_data.py:175: RuntimeWarning: overflow encountered in cast
adata.X[:, ref_idx] = X_comp
adata_arcsinh = pm.tl.normalize_arcsinh(adata, cofactor=150, inplace=False)
adata_biexp = pm.tl.normalize_biExp(adata, inplace=False)
adata_logicle = pm.tl.normalize_logicle(adata, inplace=False)
/home/runner/work/pytometry/pytometry/.nox/build-3-9/lib/python3.9/site-packages/pytometry/tools/_normalization.py:166: RuntimeWarning: invalid value encountered in scalar subtract
y = (ae2bx + p["f"]) - (ce2mdx + value)
Save data to HDF5 file format.
adata.write("example.h5ad")